NanoSIMS publications, gallery & webinar
2024
24. Annika Vaksmaa, Hortense Vielfaure, Lubos Polerecky, Michiel V.M. Kienhuis, Marcel T.J. van der Meer, Theresa Pflüger, Matthias Egger, Helge Niemann. Biodegradation of polyethylene by the marine fungus Parengyodontium album. Science of The Total Environment. doi: 10.1016/j.scitotenv.2024.172819
23. Bent Smets, Henricus T. S. Boschker, Maxwell T. Wetherington, Gérald Lelong, Silvia Hidalgo-Martinez, Lubos Polerecky, Gert Nuyts, Karolien De Wael, Filip J. R. Meysman. Multi-wavelength Raman microscopy of nickel-based electron transport in cable bacteria. Frontiers in Microbiology. doi: 10.3389/fmicb.2024.1208033
2023
22. Sofia Spataro, Bohumil Maco, Stephane Escrig, Louise Jensen, Lubos Polerecky, Graham Knott, Anders Meibom, Bernard Schneider. Stable isotope labeling and ultra-high-resolution NanoSIMS imaging reveal alpha-synuclein-induced changes in neuronal metabolism in vivo. Acta Neuropathologica Communications. doi: 10.1186/s40478-023-01608-8
21. Annika Vaksmaa, Lubos Polerecky, Nina Dombrowski, Michiel V. M. Kienhuis, Ilsa Posthuma, Jan Gerritse, Teun Boekhout, and Helge Niemann. Polyethylene degradation and assimilation by the marine yeast Rhodotorula mucilaginosa. ISME Communications. doi: 10.1038/s43705-023-00267-z
2022
20. Rob J. M. van Spanning, Qingtian Guan, Chrats Melkonian, James Gallant, Lubos Polerecky, Jean-François Flot, Bernd W. Brandt, Martin Braster, Paul Iturbe Espinoza, Joost W.Aerts, Marion M. Meima-Franke, Sander R. Piersma, Catalin M. Bunduc, Roy Ummels, Arnab Pain, Emily J. Fleming, Nicole N. van der Wel, Vasile D. Gherman, Serban M. Sarbu, Paul L. E. Bodelier, and Wilbert Bitter. Methanotrophy by a Mycobacterium species that dominates a cave microbial ecosystem. Nature Microbiology. doi: 10.1038/s41564-022-01252-3
19. S.P. Akse, L. Polerecky, M.V.M. Kienhuis, J.J. Middelburg. The influence of sediment diagenesis and aluminium on oxygen isotope exchange of diatom frustules. Geochimica et Cosmochimica Acta. doi: 10.1016/j.gca.2022.07.015
18. E. Geerken, L. de Nooijer, T. Toyofuku, A. Roepert, J.J. Middelburg, M.V.M. Kienhuis, Y. Nagai, L. Polerecky, G.-J. Reichart. High precipitation rates characterize biomineralization in the benthic foraminifer Ammonia beccarii. Geochimica et Cosmochimica Acta. doi: 10.1016/j.gca.2021.11.026
17. Nicole MJ Geerlings, Michiel VM Kienhuis, Silvia Hidalgo-Martinez, Renee Hageman, Diana Vasquez-Cardenas, Jack J Middelburg, Filip JR Meysman, Lubos Polerecky. Polyphosphate Dynamics in Cable Bacteria. Frontiers in Microbiology. doi: 10.3389/fmicb.2022.883807
2021
16. Henricus TS Boschker, Perran LM Cook, Lubos Polerecky, Raghavendran Thiruvallur Eachambadi, Helena Lozano, Silvia Hidalgo-Martinez, Dmitry Khalenkow, Valentina Spampinato, Nathalie Claes, Paromita Kundu, Da Wang, Sara Bals, Karina K Sand, Francesca Cavezza, Tom Hauffman, Jesper Tataru Bjerg, Andre G Skirtach, Kamila Kochan, Merrilyn McKee, Bayden Wood, Diana Bedolla, Alessandra Gianoncelli, Nicole MJ Geerlings, Nani Van Gerven, Han Remaut, Jeanine S Geelhoed, Ruben Millan-Solsona, Laura Fumagalli, Lars Peter Nielsen, Alexis Franquet, Jean V Manca, Gabriel Gomila, Filip JR Meysman. Efficient long-range conduction in cable bacteria through nickel protein wires. Nature communications. doi: 10.1038/s41467-021-24312-4
15. Sigrid van Grinsven, Jaap S Sinninghe Damste, John Harrison, Lubos Polerecky, Laura Villanueva. Nitrate promotes the transfer of methane‐derived carbon from the methanotroph Methylobacter sp. to the methylotroph Methylotenera sp. in eutrophic lake water. Limnology and oceanography. doi: 10.1002/lno.11648
14. Lubos Polerecky, Takako Masuda, Meri Eichner, Sophie Rabouille, Marie Vancová, Michiel VM Kienhuis, Gabor Bernát, Jose Bonomi-Barufi, Douglas Andrew Campbell, Pascal Claquin, Jan Cervený, Mario Giordano, Eva Kotabová, Jacco Kromkamp, Ana Teresa Lombardi, Martin Lukeš, Ondrej Prášil, Susanne Stephan, David Suggett, Tomas Zavřel, Kimberly H Halsey. Temporal patterns and intra- and inter-cellular variability in carbon and nitrogen assimilation by the unicellular cyanobacterium Cyanothece sp. ATCC 51142. Frontiers in Microbiology. doi: 10.3389/fmicb.2021.620915
13. Shaun P Akse, Gobind Das, Susana Agustí, Laetitia Pichevin, Lubos Polerecky, Jack J Middelburg. Imaging of organic signals in individual fossil diatom frustules with nanoSIMS and Raman spectroscopy. Marine Chemistry. doi: 10.1016/j.marchem.2020.103906
12. Lubos Polerecky, Meri Eichner, Takako Masuda, Tomáš Zavřel, Sophie Rabouille, Douglas A Campbell, Kimberly Halsey. Calculation and interpretation of substrate assimilation rates in microbial cells based on isotopic composition data obtained by nanoSIMS. Frontiers in microbiology. doi: 10.3389/fmicb.2021.621634
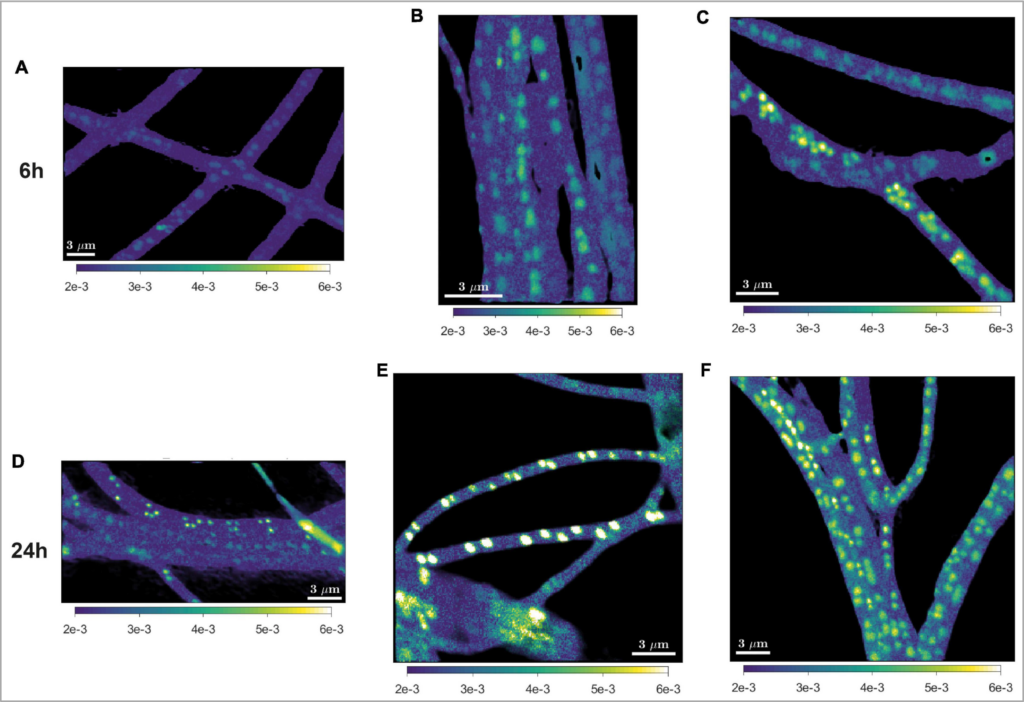
11. Nicole MJ Geerlings, Jeanine S Geelhoed, Diana Vasquez-Cardenas, Michiel VM Kienhuis, Silvia Hidalgo-Martinez, Henricus TS Boschker, Jack J Middelburg, Filip JR Meysman, Lubos Polerecky. Cell cycle, filament growth and synchronized cell division in multicellular cable bacteria. Frontiers in microbiology. doi: 10.3389/fmicb.2021.620807
2020
10. Anne Roepert, Lubos Polerecky, Esmee Geerken, Gert-Jan Reichart, Jack J Middelburg. Distribution of chlorine and fluorine in benthic foraminifera. Biogeosciences. doi: 10.5194/bg-17-4727-2020
9. Shaun P Akse, Jack J Middelburg, Helen E King, Lubos Polerecky. Rapid post-mortem oxygen isotope exchange in biogenic silica. Geochimica et Cosmochimica Acta. doi: 10.1016/j.gca.2020.06.007
8. Björn De Samber, Riet De Rycke, Michiel De Bruyne, Michiel Kienhuis, Linda Sandblad, Sylvain Bohic, Peter Cloetens, Constantin Urban, Lubos Polerecky, Laszlo Vincze. Effect of sample preparation techniques upon single cell chemical imaging: A practical comparison between synchrotron radiation based X-ray fluorescence (SR-XRF) and Nanoscopic Secondary Ion Mass Spectrometry (nano-SIMS). Analytica Chimica Acta. doi: 10.1016/j.aca.2020.01.054
7. Nicole MJ Geerlings, Cheryl Karman, Stanislav Trashin, Karel S As, Michiel VM Kienhuis, Silvia Hidalgo-Martinez, Diana Vasquez-Cardenas, Henricus TS Boschker, Karolien De Wael, Jack J Middelburg, Lubos Polerecky, Filip JR Meysman. Division of labor and growth during electrical cooperation in multicellular cable bacteria. Proceedings of the National Academy of Sciences. doi: 10.1073/pnas.1916244117
2019
6. Serginio RC Remmelzwaal, Aleksey Yu Sadekov, Ian J Parkinson, Daniela N Schmidt, Danna Titelboim, Sigal Abramovich, Anne Roepert, Michiel Kienhuis, Lubos Polerecky, Heather Goring-Harford, Katsunori Kimoto, Katherine A Allen, Kate Holland, Joseph A Stewart, Jack J Middelburg. Post-depositional overprinting of chromium in foraminifera. Earth and Planetary Science Letters. doi: 10.1016/j.epsl.2019.03.001
5. E Geerken, LJ De Nooijer, A Roepert, L Polerecky, HE King, GJ Reichart. Element banding and organic linings within chamber walls of two benthic foraminifera. Scientific reports. doi: 10.1038/s41598-019-40298-y
2018
4. Kevin J Carpenter, Maitrayee Bose, Lubos Polerecky, Alle AY Lie, Karla B Heidelberg, David A Caron. Single-Cell View of Carbon and Nitrogen Acquisition in the Mixotrophic Alga Prymnesium parvum (Haptophyta) Inferred From Stable Isotope Tracers and NanoSIMS. Frontiers in Marine Science. doi: 10.3389/fmars.2018.00157
2016
3. Wangpraseurt D, Pernice M, Guagliardo P, Kilburn MR, Clode PL, Polerecky L, Kühl M. Light microenvironment and single-cell gradients of carbon fixation in tissues of symbiont-bearing corals. ISME Journal. doi: 10.1038/ismej.2015.133
2. Fatimah Sulu-Gambari, Dorina Seitaj, Filip J. R. Meysman, Regina Schauer, Lubos Polerecky, and Caroline P. Slomp. Cable Bacteria Control Iron-Phosphorus Dynamics in Sediments of a Coastal Hypoxic Basin. Environ. Sci. Technol. doi: 10.1021/acs.est.5b04369
2015
1. Vasquez-Cardenas D, van de Vossenberg J, Polerecky L, Malkin SY, Schauer R, Hidalgo-Martinez S, Confurius V, Middelburg JJ, Meysman FJ, Boschker HT. Microbial carbon metabolism associated with electrogenic sulphur oxidation in coastal sediments. ISME Journal. doi: 10.1038/ismej.2015.10
Cyanobacteria
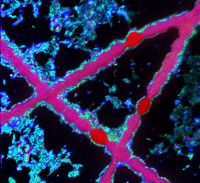
Filamentous cyanobacteria surrounded by heterotrophic bacteria. (© NanoSIMS facility UU)
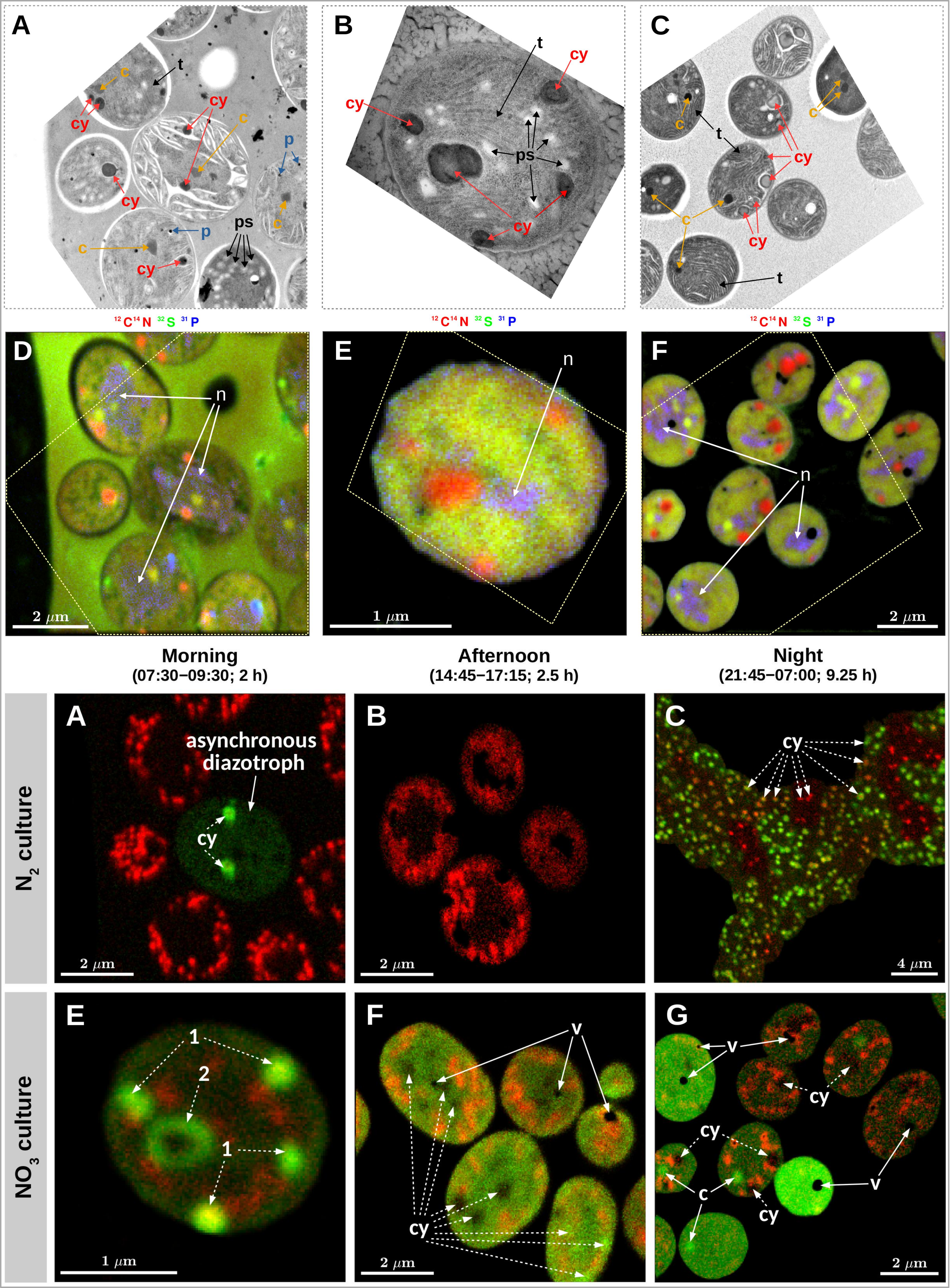
Imaging of C and N assimilation by unicellular cyanobacteria (Polerecky et al. 2021).
Cable bacteria
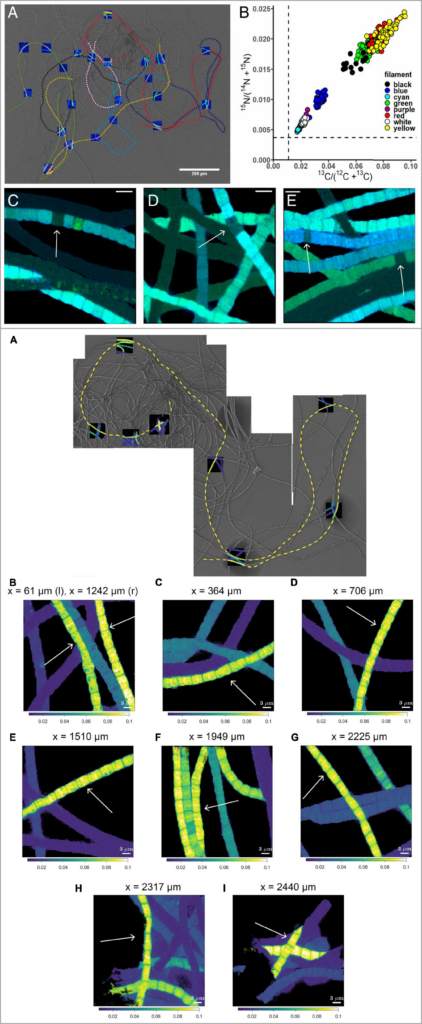
Imaging of C and N assimilation by cable bacteria (Geerlings et al. 2020and Geerlings et al. 2021).
Imaging of poly-phosphate dynamics in cable bacteria (Geerlings et al. 2022).
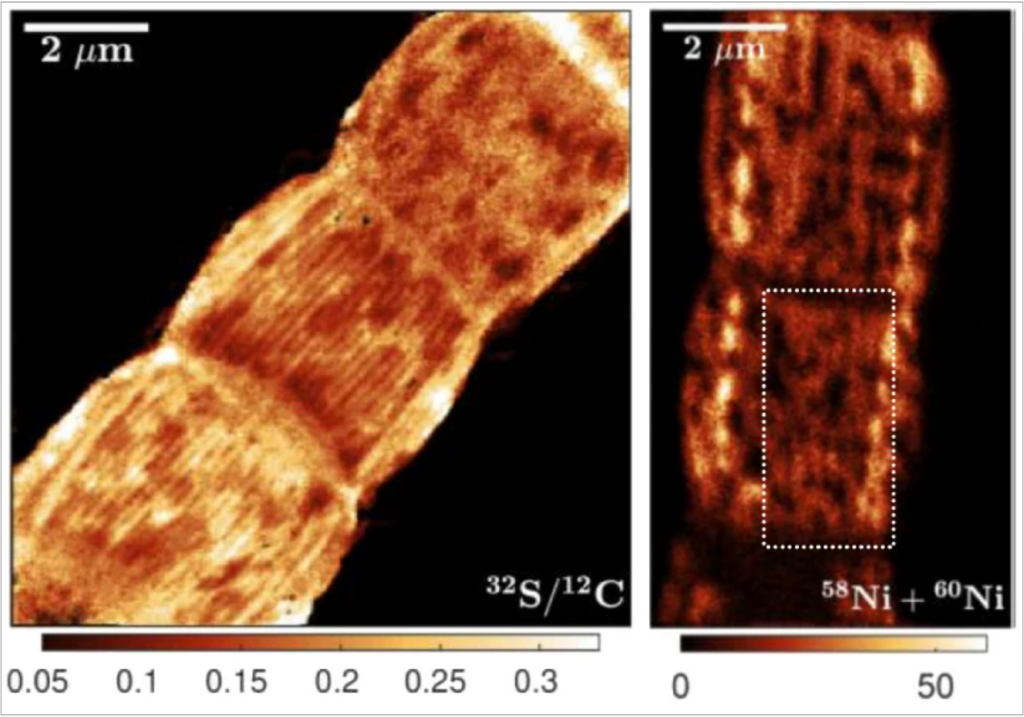
Imaging of Ni-S protein wires in cable bacteria (Boschker et al. 2021).
Diatom frustules
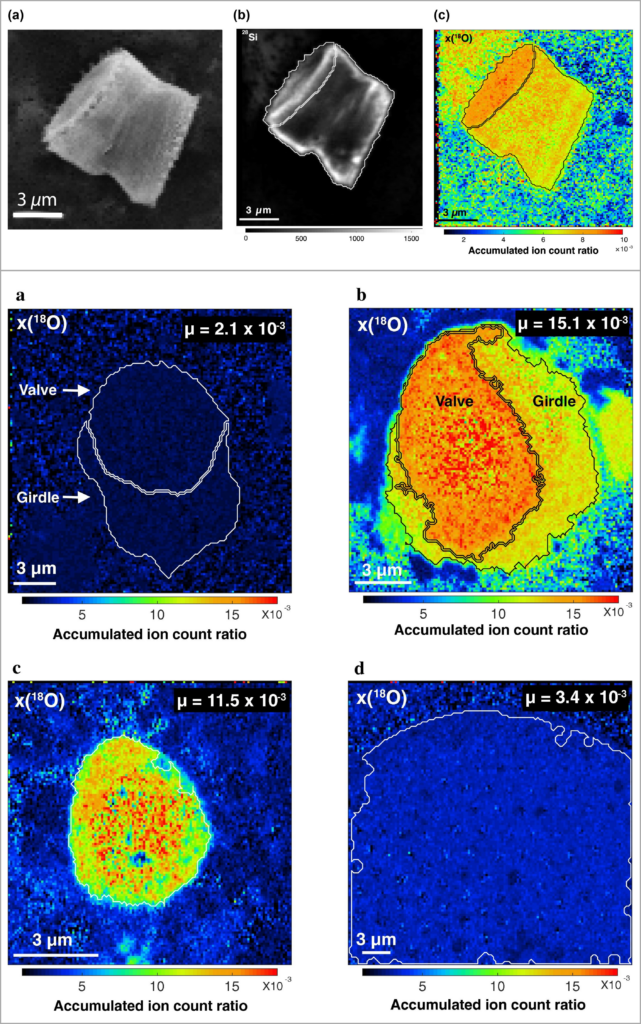
Imaging of 18O exchange between water and diatom frustules (Akse et al. 2020, Akse et al. 2022).

Imaging of organics in diatom frustules (Akse et al. 2021).
Foraminifera shells
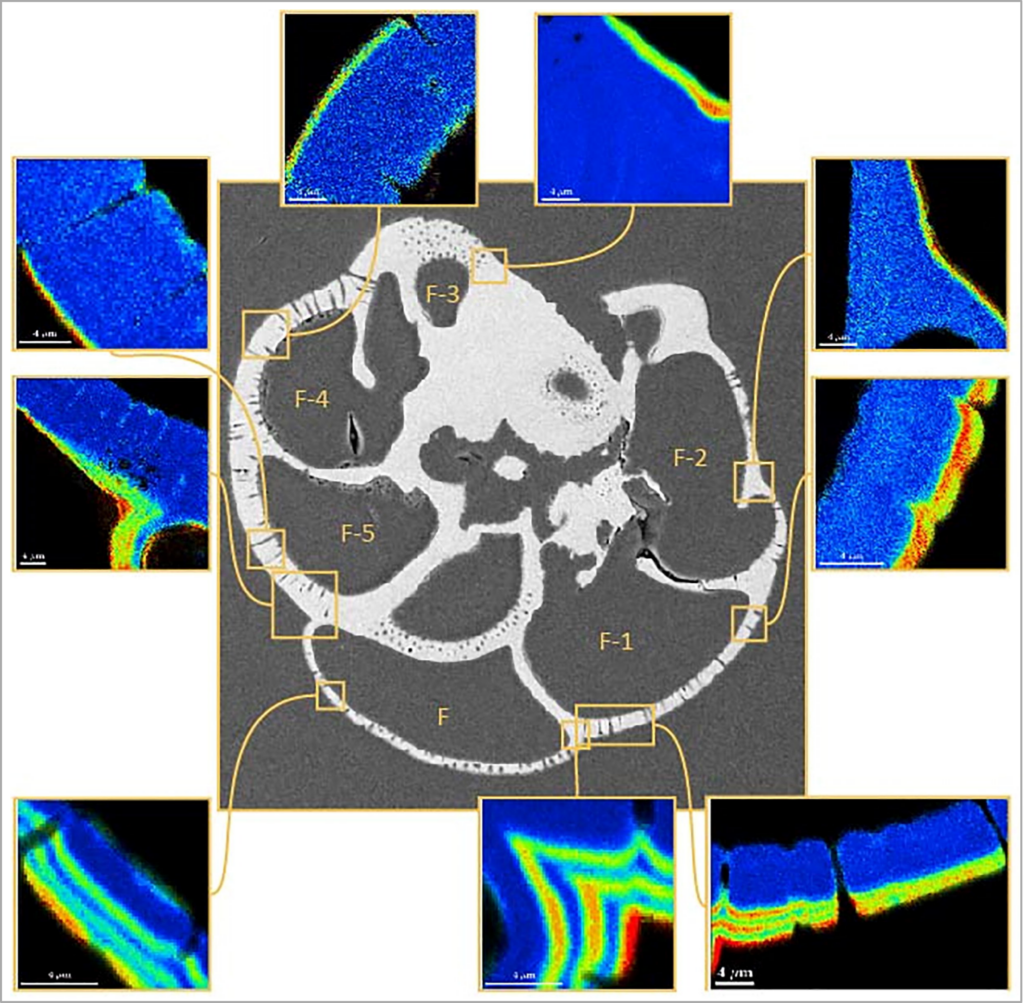
Imaging of calcite growth rates in foraminifera (Geerken et al. 2022).
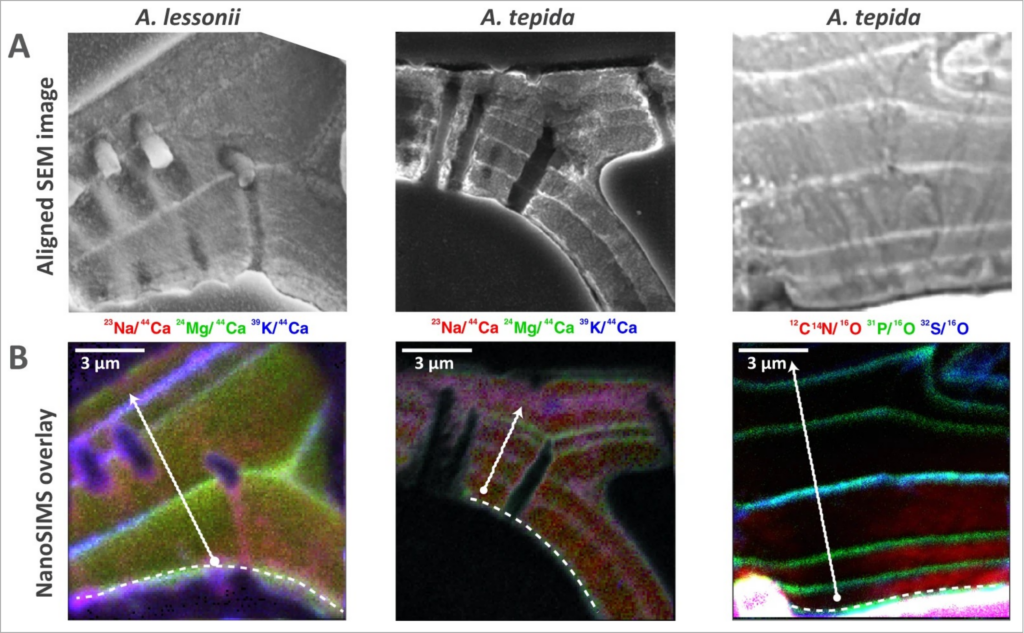
Imaging of organic linings and element banding within chamber walls of benthic foraminifera (Geerken et al. 2019).
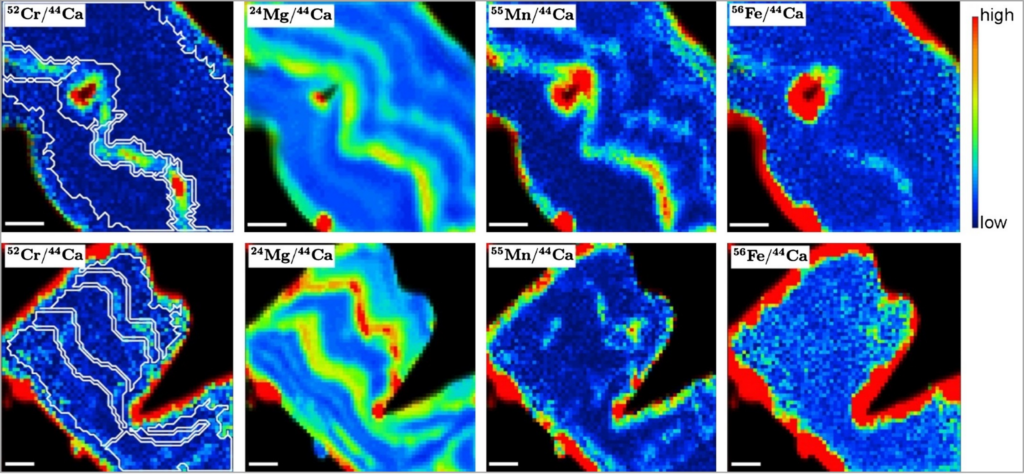
Imaging of post-depositional overprinting of chromium in foraminifera (Remmelzwaal et al. 2019).
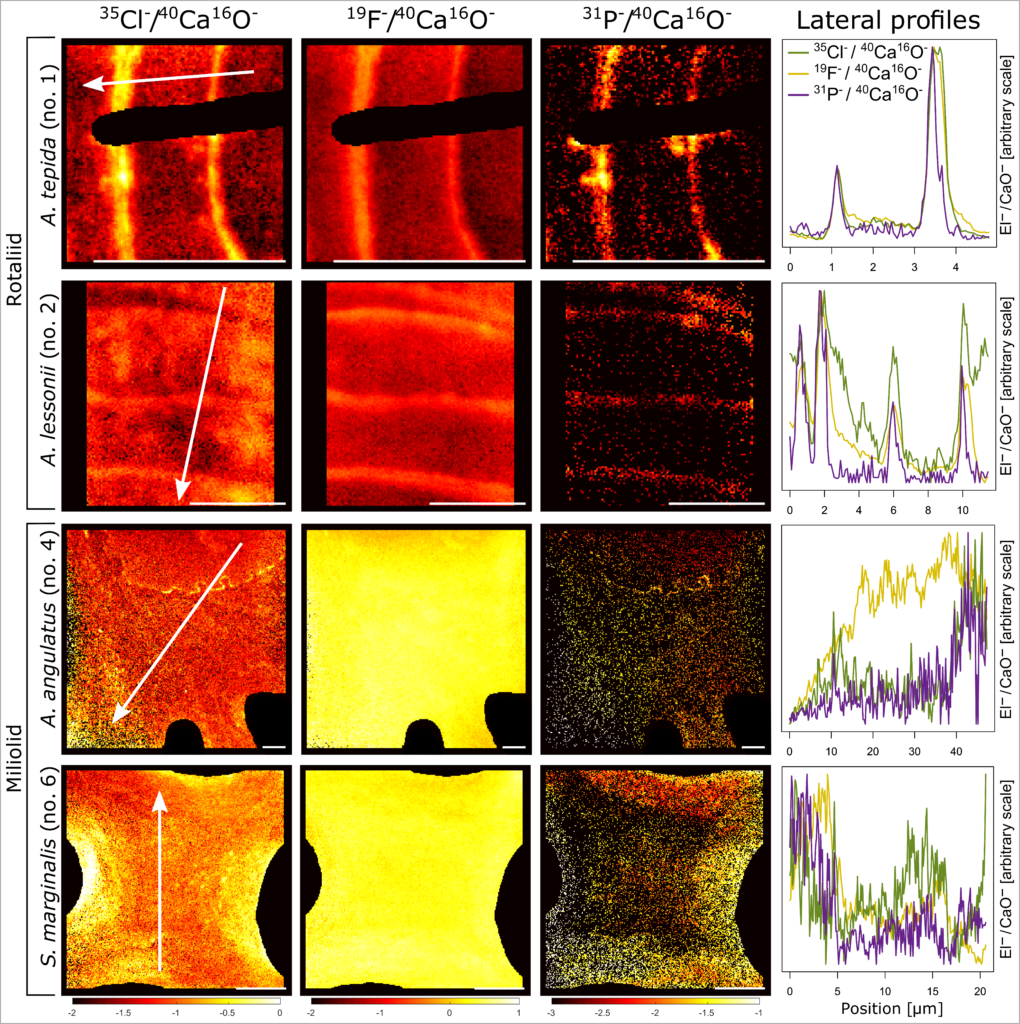
Imaging of chlorine and fluorine distributions in shells of benthic foraminifera (Roepert et al. 2020).
Methanotrophs
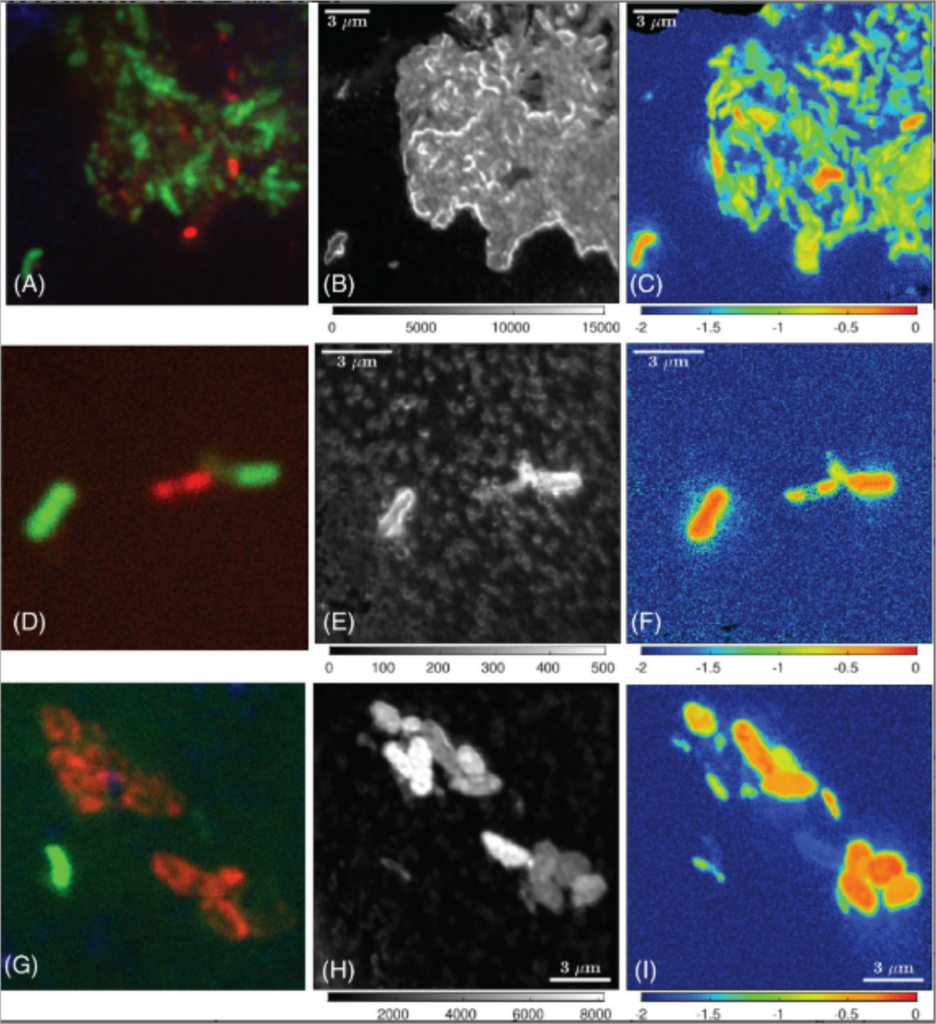
Imaging of the transfer of methane-derived C from a methanotroph to a methylotroph (van Grinsven et al. 2021).
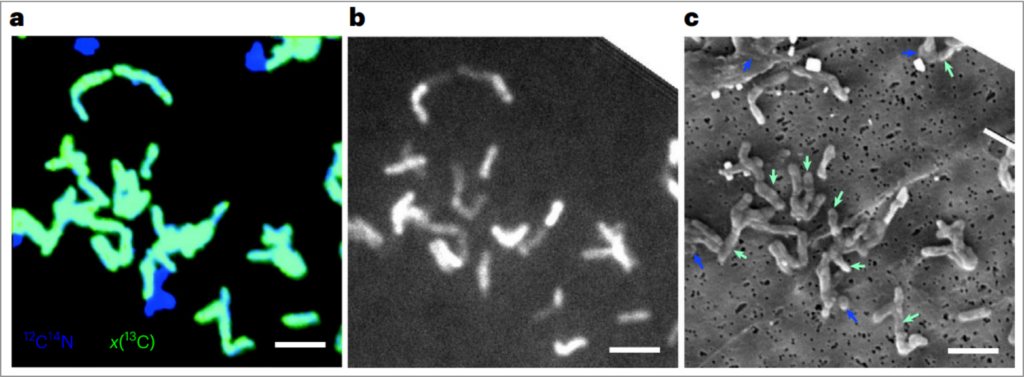
Imaging of the growth of a Mycobacterium species on methane (van Spanning et al. 2022).
Fungi
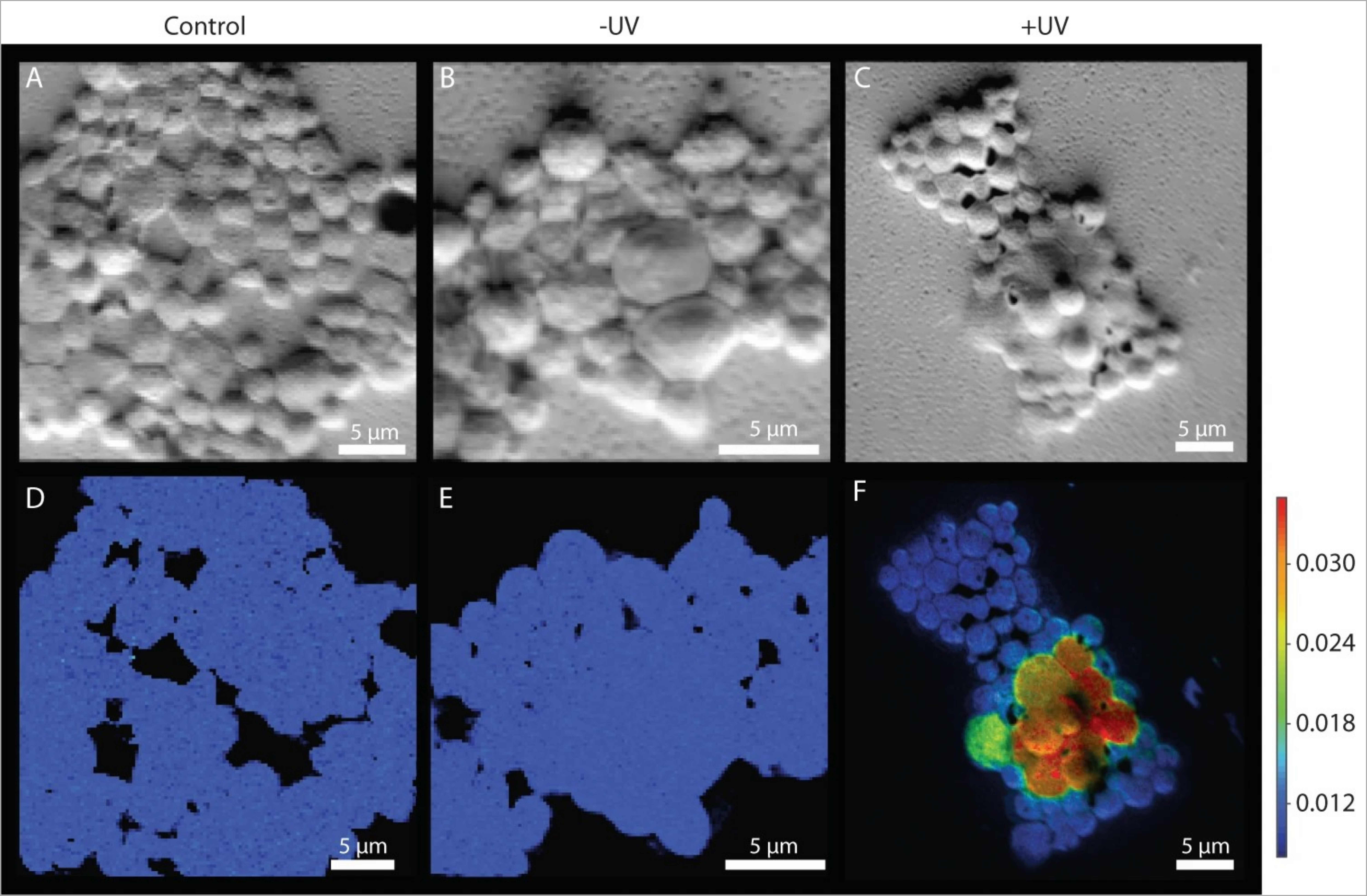
Imaging of plastic-derived carbon incorporation by marine yeast (Vaksmaa et al. 2023).
Rat models
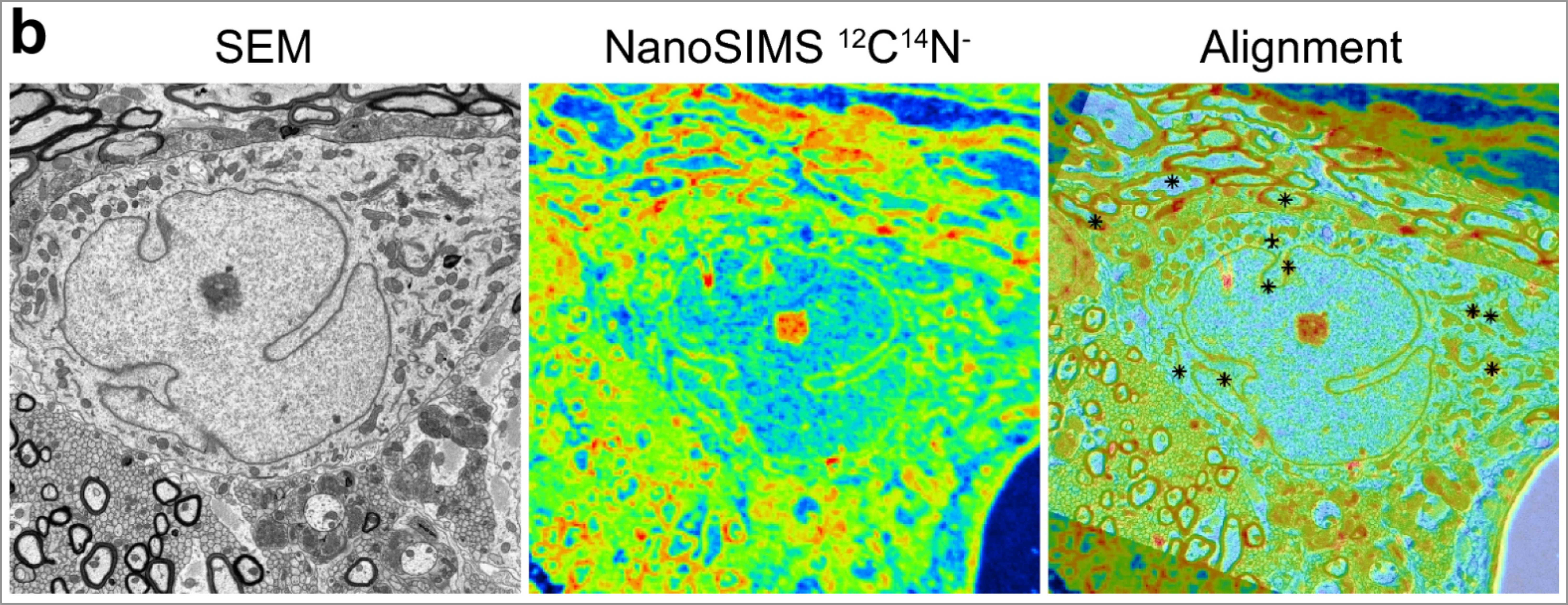
Correlative electron microscopy and NanoSIMS imaging of brain tissues in rat models (Spataro et al. 2023).
Welcome and introduction:
Use of nanoSIMS to identify and trace microbial processes and interactions
The combination of stable isotope probing (SIP) and nanoSIMS has increasingly been used in microbiology field over the past 15 years. The SIP-nanoSIMS approach is highly sensitive and allows tracing and quantifying metabolic rates at the level of single-cells, based on the assimilation of isotopically-labelled substrates into the cell biomass. It allows us to explore biological interactions between uncultured microorganisms, especially in the context of spatial and trophic relationships in biological systems where exchanges of nutrients can be extremely rapid, e.g., symbiotic or syntrophic interactions. Generally, SIP-nanoSIMS can answer questions that are out of reach for conventional approaches, ranging from microbe-host and microbe-microbe interactions (including single-cell ecophysiology) to cell-cell nutrient or metabolite exchanges, and interactions between cells and their organic/inorganic matrix. However, the wider application of this approach in microbiology field is currently hampered by the necessity of custom made, sample dependent preparation work-flows for correct identification and quantification of cellular processes and interactions within a spatial context. During this presentation, Niculina Musat (Helmholtz Centre for Environmental Research, Germany) discussed a few case studies on the application of SIP-nanoSIMS to trace microbial processes and interactions, including:
-
1. tracing the flow of carbon in the rhizosphere
2. studying the capability of microbial colonizers of plastics to degrade antibiotics
3. tracing the nutrient and water flow between fungi and microbial cells
Together, these case studies illustrate the necessity of thoughtful consideration and design of suitable sample preparation as well as the necessity of using correlative analyses to add a new level of resolution to the microbial identity-function conundrum.
Ecophysiology of multicellular cable bacteria – insights from SIP-nanoSIMS
Cable bacteria are filamentous bacteria that evolved an ingenious division of labour in which redox transformations in distant cells are coupled via long-distance electron transport. Cells in deeper sediment layers oxidize hydrogen sulfide, the electrons generated are then transported via “wires” along the longitudinal axis of the filament towards cells residing in the oxic zone, where they are used to reduce oxygen. To investigate the cellular physiology and the interactions among cells within a filament, the metabolic activity of single cells was tracked by nanoSIMS after probing with different stable isotopes (13C, 15N and 18O). Nicole Geerlings (Utrecht University) showed how these SIP-nanoSIMS analyses provided important insights into the division of labour, growth, cell cycle, and polyphosphate dynamics in multicellular cable bacteria.
Use of nanoSIMS to investigate geological processes
NanoSIMS is one of the few techniques suitable for the detection and quantification of volatiles and trace elements in natural and experimental samples at the micron scale. Laurent Remusat (National Museum of Natural History, France) presented two case to show the advances that nanoSIMS can bring to the understanding of crucial geological processes: element partitioning during planetary differentiation and crustal melting.
In 2022, we organized a webinar about the use of NanoSIMS in Life and Medical Sciences to showcase how its capabilities help us investigate biological activity on a cellular and sub-cellular level. Our aim is to stimulate future collaborations among scientists interested in this type of research. Please find the presentations below.
Welcome and introduction:
Coupling stable isotope tracer methodologies with NanoSIMS quantitative imaging
By Matthew Steinhauser (University of Pittsburg). The aim of this presentation is to present our experience coupling stable isotope tracer methodologies with NanoSIMS quantitative imaging in applications to biology and biomedical research. We have applied NanoSIMS imaging to study metabolic processes including glucose, amino acid, lipid, and nucleic acid metabolism and cell turnover in model organisms and humans at subcellular resolution. Key vignettes involving application to adipose tissue biology, cardiovascular biology, and cancer will demonstrate the breath of possibilities with this technology and underscore the power of obtaining quantitative functional measurements at the single and subcellular level.
NanoSIMS imaging of cholesterol and sphingolipids within cellular membranes
By Mary Kraft (University of Illinois Urbana-Champaign). Strategies for imaging the distributions of metabolically labeled cholesterol and sphingolipids on and within mammalian cells with a NanoSIMS instrument will be presented. By combining metabolic label incorporation and NanoSIMS imaging, the distributions cholesterol and sphingolipids in the plasma membrane and within subcellular structures within mammalian cells have been visualized. A new computational tool for converting NanoSIMS depth profiling data into accurate three-dimensional images of intracellular component distribution without requiring cell sectioning or correlated sample analysis with complementary techniques will also be presented. NanoSIMS imaging of the distributions of membrane components on and within intact cells may enable a better understanding of the roles of membrane lipids and cholesterol in cellular function.
NanoSIMS imaging of the brain in health and disease
By Silvio Rizzoli (Georg-August University, Goettingen). Brain function is intimately bound to the function of the synapses, small contact sites between neurons that enable the transfer of information. Synapse dysfunction is at the root of most brain diseases, and therefore a holistic understanding of synapse function has been the “holy grail” of neuroscience for decades. Our laboratory analyzes synapses by combining advanced imaging techniques in the optical domain with conventional mass spectrometry, with electron microscopy and with nanoscale secondary ion mass spectrometry (nanoSIMS). Over the last decade, we could derive a thorough understanding of the dynamics of synapses over long time periods, in a fashion that could not be even approached without nanoSIMS imaging, and we also approached the SIMS-based analysis of small brain-regulating drugs. We conclude that nanoSIMS is a promising tool in the biomedical domain, which has been exploited far too little.
Go back to:
→ Look@NanoSIMS software
→ Sample requirements & preparations
→ Strengths & limitations
→ NanoSIMS technique
→ Facility access & services
→ National NanoSIMS facility


